country continent year lifeExp
Afghanistan: 12 Africa :624 Min. :1952 Min. :23.60
Albania : 12 Americas:300 1st Qu.:1966 1st Qu.:48.20
Algeria : 12 Asia :396 Median :1980 Median :60.71
Angola : 12 Europe :360 Mean :1980 Mean :59.47
Argentina : 12 Oceania : 24 3rd Qu.:1993 3rd Qu.:70.85
Australia : 12 Max. :2007 Max. :82.60
(Other) :1632
pop gdpPercap
Min. :6.001e+04 Min. : 241.2
1st Qu.:2.794e+06 1st Qu.: 1202.1
Median :7.024e+06 Median : 3531.8
Mean :2.960e+07 Mean : 7215.3
3rd Qu.:1.959e+07 3rd Qu.: 9325.5
Max. :1.319e+09 Max. :113523.1
Working on nesting models following the workflow from R for Data Science.
#create year_since_1950
gapminder <- gapminder %>%
mutate(year_since_1950 = year - 1950)
#group by data that you want to produce multiple models by
#then nest to create data column of remaining columns
nest_gapminder <- gapminder %>%
group_by(continent, country) %>%
nest()
#create function for model that you want to build
#df will be the only parametre
continent_year_model <- function(df) {
lm(lifeExp ~ year_since_1950, data = df)
}
#take nested df and map function to the data column
#model will keep nested model statistics by country/year
nest_gapminder <- nest_gapminder %>%
mutate(model = map(data, continent_year_model),
glance = map(model, broom::glance))
glance_gapminder_model <- nest_gapminder %>%
unnest(glance) %>%
arrange(desc(adj.r.squared))glance_gapminder_model %>%
ggplot(.) +
geom_jitter(aes(x=continent,
y=r.squared,
color=continent)) +
ggtitle('Distribution of model fit by continent',
subtitle = 'Each dot is a country model') +
labs(x='Continent',
y='R-squared',
caption='LifeExp ~ year') +
my.theme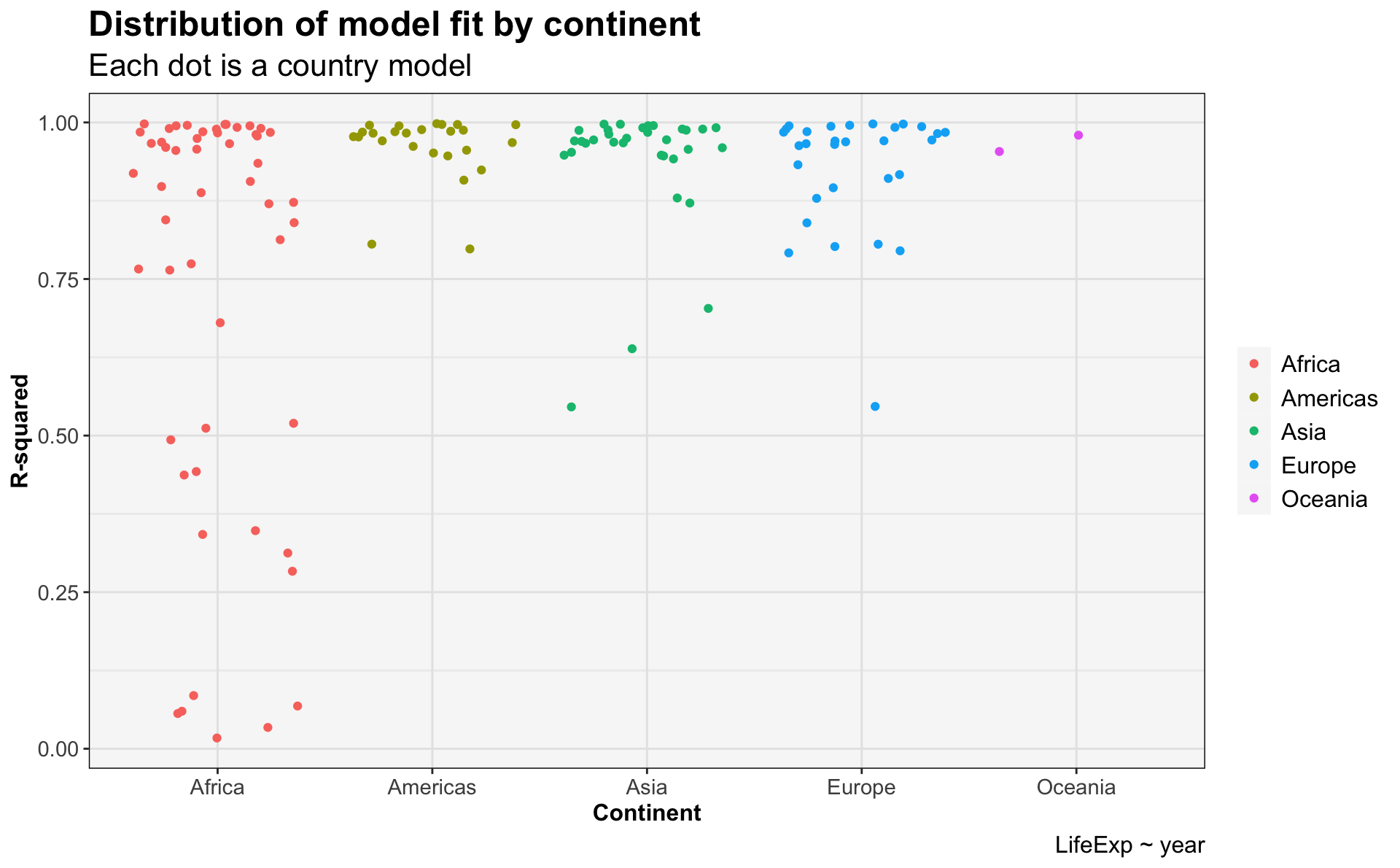
glance_gapminder_model %>%
ggplot(.) +
geom_density_ridges(aes(x=r.squared,
y=continent,
fill=continent)) +
ggtitle('Distribution of model fits by continent',
subtitle = 'Time does not explain variance in African life expectancy as strongly as others') +
labs(x='R-squared',
y='Continent',
caption='LifeExp ~ year') +
my.themePicking joint bandwidth of 0.0392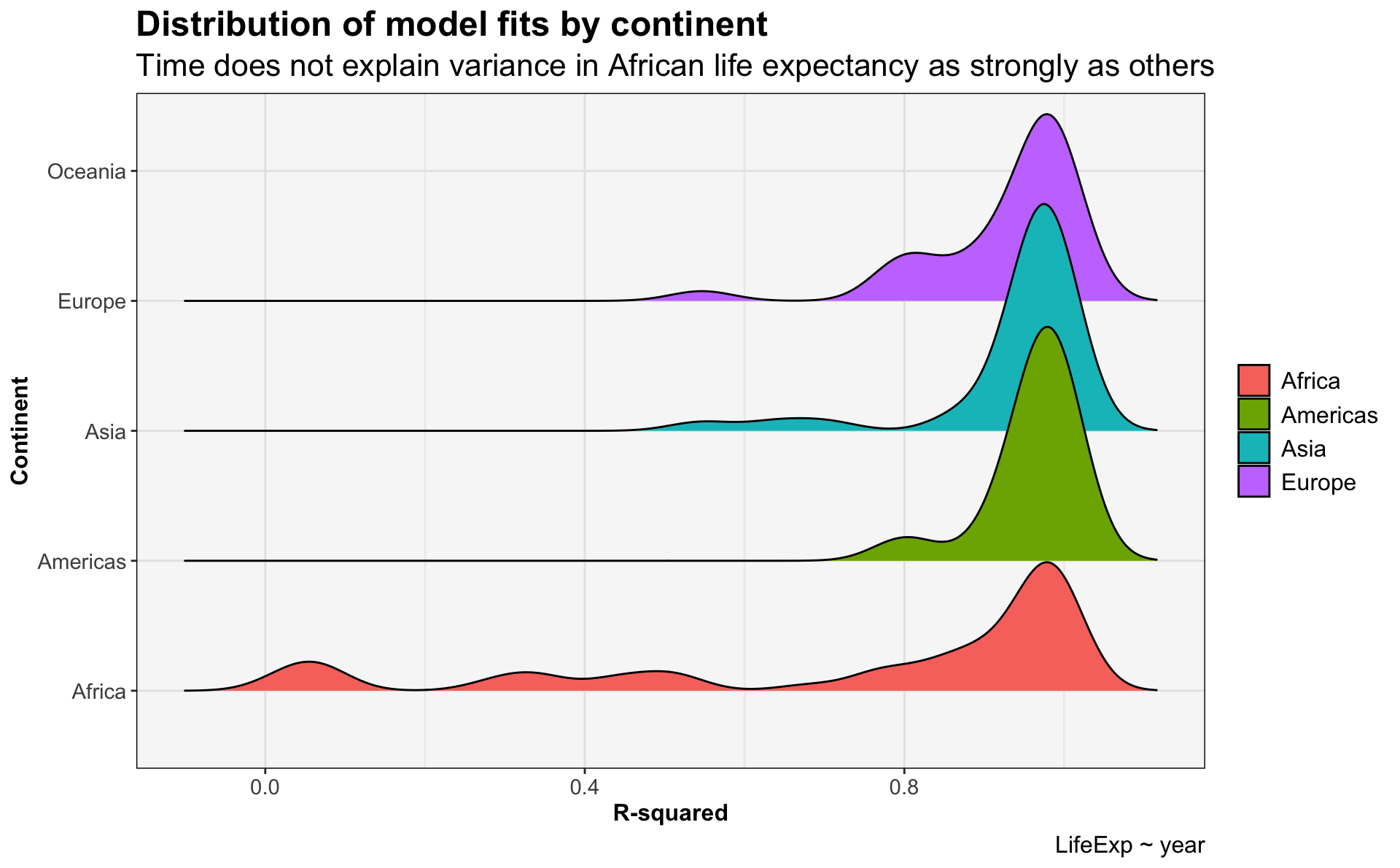
#which countries have the poor fits?
bad_fit <- glance_gapminder_model %>% filter(r.squared < 0.4)
gapminder %>%
semi_join(bad_fit, by = "country") %>%
ggplot(.) +
geom_line(aes(x=year,
y=lifeExp,
color=country), linewidth=1.1) +
ggtitle('Countries in which time does not adequately explain life expectancy',
subtitle = 'Visualizing countries with model fits less than .4') +
labs(x='Year',
y='Life expectancy',
caption='LifeExp ~ year') +
my.theme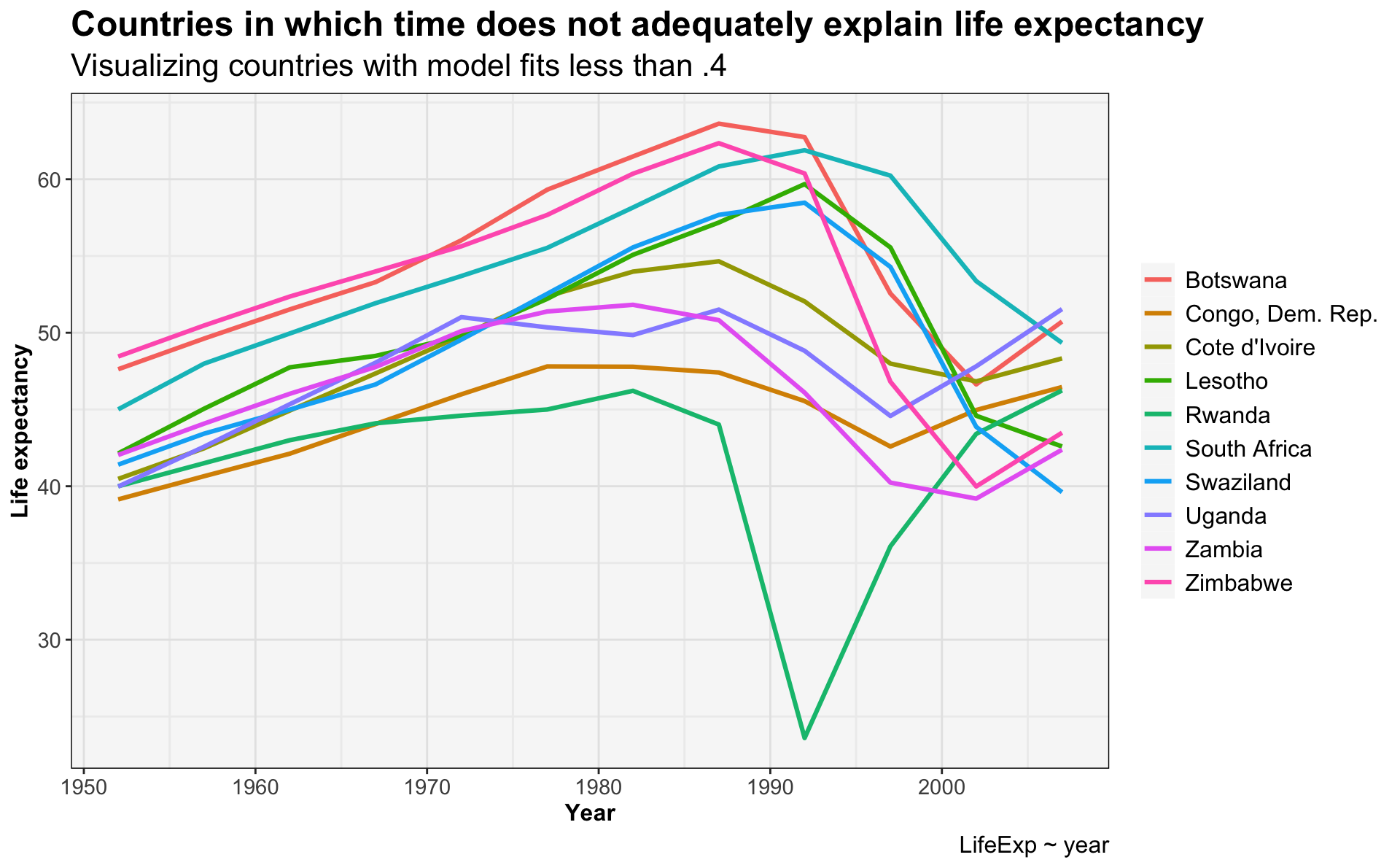
This demonstrates that:
- The 10 countries with the poorest model fits are all in Africa.
- The fit seems to be due to a dramatic decline in life expectancy that begins around 1990. This is the HIV/AIDS epidemic.
From R for Data Science (http://r4ds.had.co.nz/many-models.html#making-tidy-data-with-broom):
Making tidy data with broom:
The broom package provides three general tools for turning models into tidy data frames:
- broom::glance(model) returns a row for each model. Each column gives a model summary: either a measure of model quality, or complexity, or a combination of the two.
- broom::tidy(model) returns a row for each coefficient in the model. Each column gives information about the estimate or its variability.
- broom::augment(model, data) returns a row for each row in data, adding extra values like residuals, and influence statistics.